|
The Parkinson’s research community is currently drowning in data related to genetics. It feels like every time one comes up for air, there is a new study highlighting not one, but half a dozen novel genetic variants associated with an increased risk of developing the condition. This week alone, a new research report has been made available that by itself proposes 39 new genetic risk factors. The researchers analysed the DNA of 37,700 people with Parkinson’s and 1.4 million (!!!) healthy control subjects and found a total of 92 genetic risk factors for PD. But what does it all mean? How much influence does genetics have on Parkinson’s? In today’s post, we will outline the genetics of Parkinson’s, review some of the new studies, and discuss what the new findings mean for Parkinson’s.
|
Source: Screenrant
When I say the word ‘mutant’, what do you think of?
Perhaps your imagination drifts towards comic book superheroes or characters in movies who have acquired amazing new super powers resulting from their bodies being zapped with toxic gamma-rays or such like.
Alternatively, maybe you think of certain negative connotation associated with the word ‘mutant’. You might associate the word with terms like ‘weirdo’ or ‘oddity’, and think of the ‘freak show’ performers who used to be put on display at the travelling carnivals.
Circus freak show (photo bombing giraffe). Source: Bretlittlehales
In biology, however, the word ‘mutant’ means something utterly different.
What does ‘mutant’ mean in biology?
The word ‘mutant’ comes from the Latin mutāre which means “to change”.
In biology, we refer to a mutant when talking about small changes in DNA (which can affect the subsequent protein, which can also be referred to as a ‘mutant protein’). Many researchers prefer the words genetic variations, as the changes (or mutation) observed in DNA can vary greatly in size and nature.
Source: News-medical
It is important to understand, however, that there is nothing special about mutations.
In fact, the natural random occurrence of genetic mutations is integral to the process of evolution. And every single one of us carries genetic variations – we are each a genetic experiment. A few years back, researchers found that when parents pass their genes on to their offspring, an average of 60 new random mutations are introduced into their child’s DNA:
Title: Variation in genome-wide mutation rates within and between human families.
Authors: Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P; 1000 Genomes Project.
Journal: Nat Genet. 2011 Jun 12;43(7):712-4.
PMID: 21666693 (This report is OPEN ACCESS if you are interesting in reading it)
But what is actually meant by these small genetic variations? What exactly is a mutation in DNA?
To explain this, we need to understand a little something about basic genetics.
In almost every cell of your body you have DNA (or Deoxyribonucleic acid). It is a molecule that is composed of two chains which coil around each other, forming the famous double helix shape.
Source: Pngtree
The information stored in DNA provides the template or the instructions for making (and maintaining) a particular organism. All of the necessary details are encoded in that amazing molecule.
How is information stored in DNA?
In the image above, you can see strands reaching between the two chains of the DNA molecule, and it is these numerous strands that contain the information.
These strands are called ‘nucleotides’. These are organic molecules which contain one of four bases – the familiar A, C, T & Gs that make up our DNA code. These nucleotides form pairs (called ‘base pairs’) which then join together in long strings of DNA.

The basics of genetics. Source: CompoundChem
It takes a lot of instructions to make a fully functional human being, and as a result human DNA contains a lot of these base pairs – 3 billion of them. And having so many base pairs results in a lot of DNA in each cell – 2 meters of it. In order to get all that DNA crammed into the nucleus of a tiny little cell, DNA is wound up tightly into thread-like structures called chromosomes.
DNA in a chromosome. Source: Byjus
Humans have 23 pairs of chromosomes. And the mutations we are talking about today can occur in any of these chromosomes.
This video does a better job of explaining DNA than I do:
Now, if DNA provides the template for making a human being, it is the small variations in our individual DNA that ultimately makes each of us unique.
And these variations come in different flavours: some can simply be a single mismatched base pair (also called a point-mutation or single nucleotide polymorphism), while others are more complicated such as repeating copies of multiple base pairs.

Lots of different types of genetic variations. Source: Nature
An example of how these variation can make us who we are is red hair. The fact that an individual has red hair results from a variation in a region of DNA (called MC1R) on chromosome 16.
If that variation is not there: no red hair.
Most of the variants that we have that define who we are, we have had since conception. These are called ‘germ line’ mutations, while those that we pick up during life and that are usually specific to a particular tissue or organ in the body (such as the liver or blood), are called ‘somatic’ mutations.

Somatic vs Germline mutations. Source: AutismScienceFoundation
These variations can occur in regions of DNA that have no apparent function, and they can also appear in more functional regions (which are called genes). These genes provide regions that can be turned into RNA, which can in some cases be used to make protein.
An important aspect of these variations in genes is that you have two copies of each gene (called alleles). One copy from your father, and one copy from your mother. Call it mother nature’s insurance policy, if one copy of a gene is faulty, you have a back up. But sometimes one copy is not enough, and this can lead to problems.
In conditions that are influenced by genetics, there are two types of inherited mutations:
- A variant has to be provided by both the parents for a condition to develop – this is called an ‘autosomal recessive‘ disease
- Only one copy of the variant needs to be provided by one of the parents for a condition to develop – this is called an ‘autosomal dominant’ disease.
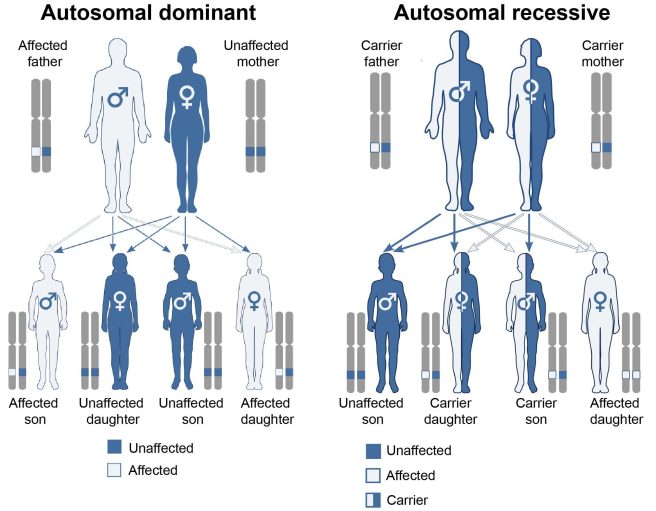
Autosomal dominant vs recessive. Source: Wikipedia
It is important to understand that most of these tiny genetic changes result in no impact on an organism. The region of DNA is not important, or biology is able to adapt and find a way around the problem.
Other variants can infer traits that may be considered beneficial, for example, some people have a genetic mutation in the CCR5 gene. HIV uses that protein to enter into human cells. Thus, if a person has a mutation in CCR5, they are extremely unlikely to become infected by the virus.
Source: Quora
There are, however, some genetic variation that are of a more serious nature – leaving the animal potentially vulnerable to developing a condition (such as BRCA1 mutations in breast cancer). And for a long time, researchers have been interested in identifying genetic variations that are associated with Parkinson’s.
EDITOR’S NOTE: Context is critical in all of these situations, as we have previously discussed in a post on the evolutionary advantage of Parkinson’s – Click here to read that post.
So what does genetics have to do with Parkinson’s?
Since it was first described by good old JP in 1812, there have been suspicions and theories of a hereditary/familial link in Parkinson’s. The condition appeared to be passed on in families – with uncles, fathers, etc also having the condition or symptoms of it.
These patterns have resulted in the idea that there could be a genetic link in Parkinson’s.
It was not until 1997, however, that the first genetic risk factor of Parkinson’s was discovered:

Title: Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease.
Authors: Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL.
Journal: Science. 1997 Jun 27;276(5321):2045-7.
PMID: 9197268
In this critical study, the researchers had encountered several independent Italian and Greek families whose members exhibited a similar early onset form of Parkinson’s. The investigators found that the affected members of those families also had genetic mutations in the long arm of chromosome 4.

The 23 pairs of chromosomes and variants (green) in chromosome 4. Source: JAMA
It was around this same time that parts of chromosome 4 was sequenced and mapped out, and as a result the researchers were able to determine that the area covering the Parkinson’s associated genetic mutations lay in a gene called SNCA, which provides the instructions for making a protein that we here at the SoPD are rather familiar with: Alpha synuclein.
You can read more about the first Parkinson’s genetic mutations in a previous SoPD post (Click here to read that post)
Over the next few years, more genetic variations that inferred a higher risk of developing Parkinson’s were identified. These discoveries gradually resulted in a list – called the PARK genes – of 20+ regions of DNA where variations were considered risk factors for developing Parkinson’s.

An early list of the PARK genes. Source: JKMA
More recently, a lot of new genetic risk factors for Parkinson’s have been identified. A good example of this is a study published last year (2017):
Title: A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci.
Authors: Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F; International Parkinson’s Disease Genomics Consortium; 23andMe Research Team, Kerchner GA, Ayalon G, Bingol B, Sheng M, Hinds D, Behrens TW, Singleton AB, Bhangale TR, Graham RR.
Journal: Nature Genet. 2017 Oct;49(10):1511-1516.
PMID: 28892059 (This report is OPEN ACCESS if you would like to read it)
In this study, the researchers conducted a GWAS comparing DNA from 6,476 people with Parkinson’s with DNA from 302,042 controls. They then compared those results with another GWAS dataset from a recent study involving 13,000 people with Parkinson’s and 95,000 controls.
Wait a minute. What is a GWAS?
A genome-wide association study (or GWAS) is an analysis of a set of genetic variants across the entire genome (or all of the DNA in your cells, including mitochondrial), and this analysis is conducted in a large pool of different individuals to see if any variants are associated with a particular trait (or medical condition). It is typically an analysis of single nucleotide polymorphisms (or SNPs; a variation in a single nucleotide). The researcher will check your DNA for the presence of a large set of single nucleotide variations, and then compare them with the results collected from other people.
Source: Knowgenetics
Now as we have discussed above, we all have these tiny mutations. But what a GWAS does, is seek to determine whether people with a particular trait (for example, constrictions in small blood vessels) have a shared single nucleotide polymorphisms, compared to people that do not have that trait. Think of the example of red hair mentioned above, a GWAS analysis of red haired people would point towards a variation on chromosome 16 (in the MC1R gene).
And the result can be presented in what is called a “Manhattan plot” – Manhattan plots get their name because they often look like the city skyline of Manhattan:
Source: Andrewprokos
Across the bottom of a Manhattan plot are the chromosomes, and each dot represents a single nucleotide polymorphisms for that particular region of the DNA. The more dots there are above that region – the more people with a variation in that region – the higher the dot in that particular location. The Manhattan plot in the image below is for an analysis of people with constrictions in small blood vessels, and what you can see is that the number of dark blue dots above chromosome 19 indicate that there are a lot of people who have this trait that also have a genetic variant in that region.
Source: Wikipedia
When researchers who conducted the Parkinson’s study from last year analysed of their data, this was the kind of Manhattan plot they observed:
Source: PMC
Immediately you can hopefully see that we are looking at a complicated result – there are a lot of ‘buildings’ in this Manhattan city skyline. Lots of genetic variants that could be considered risk factors for Parkinson’s.
The researchers then tested these results in a third data set of DNA from 5,851 people with Parkinson’s and 5,866 controls. From this combined analysis (totalling 26,035 PD cases and 403,190 controls), they found 17 novel risk factors for Parkinson’s (41 in total; 24 had been previously reported).
This is great, but what can we do with all this information?
An interesting use of this data is what it can tell us about the biology of Parkinson’s:
Title: Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease.
Authors: Robak LA, Jansen IE, van Rooij J, Uitterlinden AG, Kraaij R, Jankovic J; International Parkinson’sDisease Genomics Consortium (IPDGC), Heutink P, Shulman JM; IPDGC Consortium members; International Parkinson’s Disease Genomics Consortium (IPDGC).
Journal: Brain. 2017 Nov 13.
PMID: 29140481
In this study (also from 2017), when the researchers focused their analysis on genetic variants among 54 lysosomal storage disorder genes (these are genes involved in the process of waste disposal or recycling), they found that the majority (56%) of the Parkinson’s cases they analysed had at least one variant in a lysosomal storage disorder gene (and 21% had more than one).
The researchers concluded that multiple genetic hits may act in combination to degrade lysosomal function, which could in turn enhance Parkinson’s susceptibility (perhaps by limiting the clearance of aggregating proteins).
In medicine, a “forme fruste” (French, “crude or unfinished, form”) is the manifestation of a disease or syndrome, in an incomplete or partial state. And the researchers behind this lysosomal study asked whether some cases of Parkinson’s could be a forme fruste of lysosomal storage disease.
Source: PathologyOutlines
Now, such knowledge could be useful in determining who in the Parkinson’s community may respond better to treatments designed to boost the waste disposal system (this was the goal of the Ambroxol trial we are currently awaiting the results of).

Ambroxol. Source: Skinflint
Perhaps future clinical trials (or basic treatment) will involve firstly genetically testing participants to determine if they have genetic variants associated with a particular biological function that could be therapeutically targeted.
We are not there yet with our understanding of the biology, but this is a hypothetical possibility.
Interesting. What was the new research presented this week?
This week, researchers made a manuscript available on the pre-peer review/pre-publication, OPEN ACCESS depository website called BioRxiv.
Source: BioRxiv
I don’t like discussing research that hasn’t been through the peer-review process (he says having discussed BioRxiv manuscript 3 or 4 times in previous posts!), but in this case the study and the results are really amazing!
Here is the manuscript in question:
Title: Parkinson’s disease genetics: identifying novel risk loci, providing causal insights and improving estimates of heritable risk
Authors: Nalls MA, et al (there were a lot of researchers involved in this study), Singleton AB, The 23andMe Research Team, System Genomics of Parkinson’s Disease (SGPD) Consortium, International Parkinson’s Disease Genomics Consortium
Database: BioRxiv
DOI: https://doi.org/10.1101/388165 (This manuscript is OPEN ACCESS if you would like to read it)
In this study, the researchers had collected data related to DNA from 37,700 people with Parkinson’s as well as 1.4 million (!!!) control subjects. Their analysis involved looking at 11.4 million (!!!) single nucleotide variants.
Please stop for a moment and just think about the scale of this analysis – they collected sequenced DNA data from almost 1.5 million people and analysed each of those files for 11.4 million possible errors in the genetic code.
That is a HUGE study.
Certainly the most comprehensive genetic analysis of Parkinson’s to date.
And what did they find?
Well, the results of the analysis were also huge: This new study identified 92 genetic risk factors, of which 39 were completely novel.
Read that again: almost half of the genetic risk factors were previously unknown!
The risk factors were presented in an Iris plot, which is similar to a Manhattan plot except circular. The same principles apply with the red dots representing the chromosomal location of major genetic risk factors. For example, in the image below there are a lot of red dots over the 4th chromosome, which is where the alpha synuclein gene is located:
An Iris plot of the results (red dots indicate location of major risk factors). Source: BioRxiv
The results provide the research community with a great deal of new genes (and biological pathways) to investigate in the context of Parkinson’s.
And the researchers believe that a considerable genetic component of Parkinson’s is still yet to be discovered. They seem very keen to expand on this huge data set. As they wrote in their discussion of the results: “Put simply, more samples, more data, more understanding, and more potential”.
These current results allow the researchers to explain between 11-15% of Parkinson’s genetic burden. And while this may seem like a low number, this kind of percentage is actually becoming quite common in the study of genetics. Curiously, GWAS research is having a hard time explaining traits that are largely assumed to be determined by genetics, for example ‘height’:

Title: Rare and low-frequency coding variants alter human adult height
Authors: Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C,………at least 200 additional authors have been deleted here in order to save some space…….EPIC-InterAct Consortium; CHD Exome+ Consortium; ExomeBP Consortium; T2D-Genes Consortium; GoT2D Genes Consortium; Global Lipids Genetics Consortium; ReproGen Consortium; MAGIC Investigators, Rotter JI, Boehnke M, Kathiresan S, McCarthy MI, Willer CJ, Stefansson K, Borecki IB, Liu DJ, North KE, Heard-Costa NL, Pers TH, Lindgren CM, Oxvig C, Kutalik Z, Rivadeneira F, Loos RJ, Frayling TM, Hirschhorn JN, Deloukas P, Lettre G.
Journal: Nature. 2017 Feb 9;542(7640):186-190.
PMID: 28146470
In this study, the researchers – who are part of the GIANT consortium – were analysing DNA collected from over 700,000 people and trying to determine which genetic variants could influence height.
I’ll let you decide if height is important for acting. Source: Vulture
Why study height?
Good question. There are several reasons:
- it is extremely easy to accurately measure (think tape measure)
- the researchers believed that if we can master the complex genetics of something simple like height maybe what we learn will give us a blueprint for how we should study more complex medical disorders.
Ok, so what did they find?
Well,… in a previous report published in 2014, the researchers had used DNA from 253,288 individuals and discovered 697 genetic variations that appear to influence height (Click here to read an OPEN ACCESS version of that report). Each of those genetic variants had only a tiny impact on the height of an individual (increasing it by mere millimetres), and collectively they could only explain about 16 percent of the variation in height (for people of European ancestry at least).
In their more recent GIANT report, the same investigators reported that they have now analysed DNA collected from over 700,000 people and that they had found 83 additional genetic variants. Interestingly, 24 of these newly discovered genetic variants affect height by more than 1 cm (4/10 of an inch), but they are extremely rare genetic differences that only becoming apparent in this larger data set.
And with these new findings, approximately 27.4 percent of the heritability of height can now be accounted for by genetics.
Which begs the question you are probably pondering….
What about the other 72.6%?
Another good question.
And the answer is really simple:
We don’t know.
And it will be hard working it out as previous estimations based on family and twin studies have suggested that genetics accounts for at least 80% of height (Click here to read more about this). Thus, we are missing a huge chunk of the picture.
And this may also be the same case in Parkinson’s. Except…
Except what?
Well, height is a relatively simple thing to investigate. You grab your tape measure and politely ask everyone to line up to be measured. We are definitely not be looking at a simple situation in the case of Parkinson’s. With Parkinson’s, we may be dealing with a bunch of conditions that give rise to a similar physical appearance (rigidity, slowness and tremor), but completely different underlying biology. And some of those conditions could have multiple underlying genetic risk factors, while others may be more influenced by yet to be determined environmental factors.
There is still a great deal to be learned.
So what does it all mean?
Genetic variations have played an important role in not only making us humans, but also individuals. These variations are part of a process of trial and error, that over billions of years has resulted in marvellous biological structures (such as the eye ball, the eldow, the tongue, and the brain) and in amazing creatures such as Platypus (did you know, that platypus have electrolocation – they can sense the electrical signals in their prey’s muscles).
A moderate frequency of family history of Parkinson’s has lead to the belief that there is a certain level of genetic influence in the condition. Only recently, however, has technology arrived at a point where the means of analysis required for dealing with this issue is available.
This week researchers made available a new report, which outlines a MASSIVE analysis of the genetics of Parkinson’s. DNA from 37,000 people with PD has been used in the analysis and it has highlighted new genetic risk factors for this condition. As we have discussed above, this is potentially a treasure trove of new information for researchers – numerous new biological pathways to be investigated – but it also helps to the Parkinson’s community to build a view of how this condition could be treated in the future.
New therapeutic agents – some of which are currently being clinically tested – are specifically focused on some of the biological pathways that have been determined by previous genetic analysis (for example LRRK2 – Click here to read a recent post on this topic). And further understanding of these pathways will hopefully lead to better therapies and more personalised treatment.
EDITOR’S NOTE: The new manuscript that has been discussed in this post has not yet been through the peer-review process of publication. If the final published article changes significantly from the one presented here, we will adapt this post accordingly.
The banner for today’s post was sourced from Reallifecam





















My family on both sides don’t have any history of Parkinson’s So how did I end up with it. ?
LikeLike
Hi Pete,
Thanks for your comment.
This is a question that many in the community are obviously asking. If less than 20% of Parkinson’s cases can be explained by genetics, what has caused the rest of them. Environmental factors are suspected of being involved – from pesticides to viruses. But nailing down which of them is the major influencer, is proving to be a difficult task. I guess if it was a simple problem, we probably would have answered it by now.
Sorry I can’t provide a better answer.
Kind regards,
Simon
LikeLike
Simon, thank you for another excellent post that explains progress in research of genetic aspect of PD. It was not clear to me if the following statement is true: if you happen to have 2 copies of a particular PD related mutation it means that you WILL develop PD. If you have only one copy you MIGHT develop PD. Also I wonder if all of these genes are unique to humans only or if other species have them as well. If other animals have them and similar mutations are found there too one would expect to find PD in those species but if I understand it right PD is uniquely human.
Thanks,
Felix
LikeLike
Hi Felix,
Thanks for your comment – and for the interesting question.
You can have multiple copies of a genetic risk factor for Parkinson’s and not develop the condition. This is where Parkinson’s differs from Huntington’s disease. With Huntington’s, if you have the genetic variant you are definitely going to develop Huntington’s. But in Parkinson’s, we have individuals carrying genetic risk factors who do not develop the condition. For example, there is a lady in Australia with two copies of a PARKIN mutation who has not developed Parkinson’s (now in her 80s), while her daughter has developed a very early onset version of PD (https://scienceofparkinsons.com/2017/10/07/nix/).
As far as I’m aware, most of these variants are unique to humans (and I’d be happy to be corrected on this). And even when we engineer mice to have these genetic variants, they do not develop the features of PD. Until recently, we have had very few genetic models of PD. Recently, researchers analysed differences in the Parkinson’s-associated protein alpha synuclein between different types of animals. They collected alpha synuclein from elephants, bowhead whales, & pigs, and then compared it with the human protein (https://pubs.acs.org/doi/10.1021/acs.biochem.8b00627). They found some interesting differences between species, some of which appeared to make the other species versions of alpha synuclein less likely to cluster together.
The genetics of PD is a real mystery – fascinating, but it would be nice to have some answers.
I hope this has at least answered your question.
Kind regards,
Simon
LikeLike
Simon,
Thank you for this response. In one of your posts you mentioned that there was study done recently that found virus like particles in samples of post-mortem collected brain tissues of PwP. You probably referenced this publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5968367/ . Although the number of samples was quite small the authors make a pretty convincing case for the viral theory of PD pathogenesis. I wonder if genetic mutations affect immune response to the virus infection (assuming virus is the actual trigger for the onset of PD) thus making PD is more likely in some individuals. I am also curious why viral theory of PD to this day didn’t get solid substantiation (referenced here publication mentions few facts however it also mentioned others that would make this theory very plausible).
LikeLike