|
# # # # RNA – the usable copy of a section of DNA – has regions called introns that need to be removed before the RNA can be used for the production of protein. The process of removing introns is called splicing. Recently researchers have noticed that a genetic mutation in a Parkinson’s-associated gene – called DJ-1 – affects the splicing of the associated RNA and this has serious consequences on the activity of the DJ-1 protein. Interestingly, they were able to pharmacologically rescue the effect, and noticed that DJ-1 might not be the only Parkinson’s-associated gene affected by this splicing error. In today’s post, we will discuss what splicing is, review the new research, and discuss the wider implications for the Parkinson’s community. # # # # |
 Source: Wikibooks
Source: WikibooksToday’s post starts off with a definition:
Splice – /splʌɪs/; verb;
Meaning: “to combine, interweave”.
Origin: 16th century: probably from Middle Dutchsplissen,
Similar: braid, plait, entwine, intertwine, interlace, knit
Additional/alternative meanings:
1. (From the arts) When two pieces of recorded music – with a similar key and tempo – are combined:
2. (From biology) The process that removes the intervening, non-coding sequences of genes (introns) from pre-mRNA and joins the protein-coding sequences (exons) together in order to enable translation of mRNA into a protein:
Ok, so the first alternative definition about music I understood and the video was helpful, but can you explain the second definition in more detail please?
Sure.
You may recall from your high school science class, the teacher talking about the central dogma of biology: Deoxyribonucleic acid (or DNA) gives rise to Ribonucleic acid (or RNA), and RNA gives rise to protein.
That is one of the keys to all of life as we know it: DNA begets RNA, which begets protein.

The basic of biology. Source: Youtube
DNA provides the instructions for making a protein, RNA – sometimes called messenger RNA (or mRNA) – is the template (or a photocopy of the designs) for making a protein, and protein is generally considered the functional end product in the process.
It is slightly more complicated than that though.
Not all of your DNA can be copied into RNA. There are specific regions of DNA that are referred to as genes. And it’s these genes that are copied into RNA. The process of “copying” DNA into RNA is called transcription, and this occurs inside the nucleus of a cell – where the DNA is safely stored.
 Source: Expii
Source: Expii
The process of RNA being used to produce protein is called translation, and this occurs outside of the nucleus in a structure called the endoplasmic reticulum . But this is a very simplistic explanation of an extremely complicated set of processes that are required for making a protein. A lot of small, but critical steps are required.
An example of a critical step in translation is splicing.
When RNA is transcribed from DNA, it is not ready to be used for making protein. The newly formed RNA – which is referred to as pre-mRNA – contains pieces that need to be removed before it can be used in translation.
Each gene and subsequent pre-mRNA contains exons and introns.
Exons are sections of a gene that are included in mature RNA. Introns are regions of a gene that need to be removed from a piece of RNA before it can be used to make protein. The process of removing introns is called “splicing”.
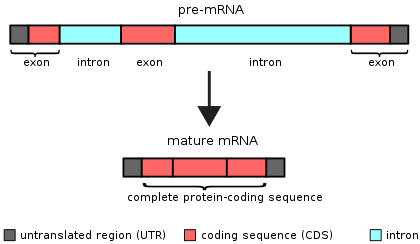 Source: Wikipedia
Source: Wikipedia
The removal of introns involves the spliceosome (cool name!). This is a complex of proteins (called snRNPs – small nuclear ribonucleo proteins, pronounced “snurps”), which work together to cut the intron out and join the loose ends of the exons.
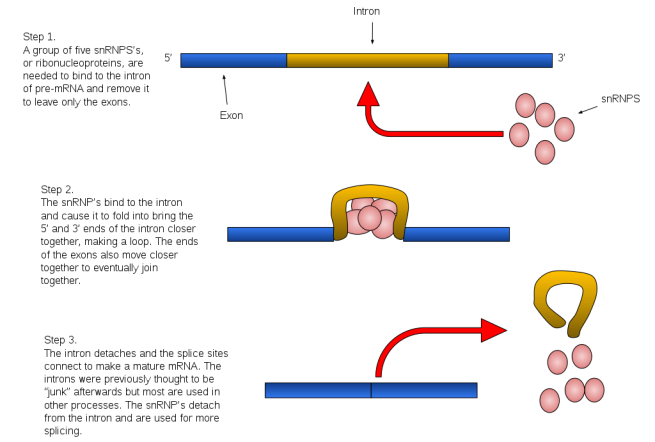 Source: Wikipedia
Source: Wikipedia
The spliceosome is primarily composed of five snRNPs (termed U1, U2, U4, U5, and U6), but it also forms different complexes during the splicing process, each serving different aspects of the process.
Think of the process of splicing as a room of film editors, who selectively cut out section of unnecessary film (the introns) from the 3-hour long ‘director’s cut’ version of a film (the pre mRNA) to be able to send a shorter, more cleaned-up final cut version (mature mRNA) to the cinemas for distribution.
What happens when something goes wrong with splicing?
Good question. This is what the rest of this post is about.
Curiously, it has been estimated that 1/3 of all disease-causing genetic mutations impact on splicing (Click here to read more about this). Genetic mutations of splicing sites can result in loss of function of necessary proteins, causing cellular dysfunction.
|
# RECAP #1: RNA contains introns and exons when it is first produced. The introns need to be removed before the RNA can be used to make protein (“translation”). The removal of introns is called splicing. # |
Interesting. So what does splicing have to do with Parkinson’s?
Recently this report was published:
 Title: A patient-based model of RNA mis-splicing uncovers treatment targets in Parkinson’s disease.
Title: A patient-based model of RNA mis-splicing uncovers treatment targets in Parkinson’s disease.
Authors: Boussaad I, Obermaier CD, Hanss Z, Bobbili DR, Bolognin S, Glaab E, Wołyńska K, Weisschuh N, De Conti L, May C, Giesert F, Grossmann D, Lambert A, Kirchen S, Biryukov M, Burbulla LF, Massart F, Bohler J, Cruciani G, Schmid B, Kurz-Drexler A, May P, Duga S, Klein C, Schwamborn JC, Marcus K, Woitalla D, Vogt Weisenhorn DM, Wurst W, Baralle M, Krainc D, Gasser T, Wissinger B, Krüger R.
Journal: Sci Transl Med. 2020 Sep 9;12(560):eaau3960.
PMID: 32908004
In this study the researchers were interested in a Parkinson’s associated protein called DJ-1.
What is DJ-1?
Back in 2003, this research report was published:
 Title: Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism
Title: Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism
Authors: Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P.
Journal: Science. 2003 Jan 10;299(5604):256-9.
PMID: 12446870
In this study, the researchers had been presented with two families from genetically isolated communities (in the Netherlands and Italy) that had members displaying an early-onset form of Parkinson’s. They had previously identified a region of DNA on chromosome 1 that they called PARK7 (Click here to read that report), and they had also validated that result on an independent database of DNA from people with early-onset Parkinson’s (Click here to read more about that study). But it was not until 2003 that they identified the specific gene within the PARK7 region on chromosome 1.
And that gene was DJ-1.
The researchers immediately started looking for more cases young-onset Parkinson’s to see how common the DJ-1 mutation was. And that effort resulted in a research report that was published later that same year:
 Title: Early-onset Parkinson’s disease caused by a compound heterozygous DJ-1 mutation.
Title: Early-onset Parkinson’s disease caused by a compound heterozygous DJ-1 mutation.
Authors: Hague S, Rogaeva E, Hernandez D, Gulick C, Singleton A, Hanson M, Johnson J, Weiser R, Gallardo M, Ravina B, Gwinn-Hardy K, Crawley A, St George-Hyslop PH, Lang AE, Heutink P, Bonifati V, Hardy J, Singleton A.
Journal: Ann Neurol. 2003 Aug;54(2):271-4.
PMID: 12891685
In this study, the investigators collected DNA from 107 early-onset (that is age at diagnosis being below 50 years of age) Parkinson’s and conducted DNA sequencing on the entire DJ-1 gene. They identified several different mutations within the DJ-1 gene that were shared between independent subjects in their 107 cohort.
With subsequent research, investigators have identified more than 25 mutations within the PARK7 region of DNA that are associated with early-onset form of Parkinson’s. Some PARK7 gene mutations lead to an abnormally small DJ-1 protein which is unstable and does not function properly. Other mutations delete a large portion of the PARK7 gene which prevents the production of any functional DJ-1 protein.
What does DJ-1 protein do?
DJ-1 is present in almost all cells and it is a very busy protein, involved in many different roles required for normal biological activity. One of the primary functions of DJ-1 is to help protect cells from oxidative stress.
What is oxidative stress?
Oxidative stress is the damage done to a cell by the process of oxidation.
And yes, I know what you are going to ask next: What is oxidation?
Oxidation is the loss of electrons from a molecule, which in turn destabilises that particular molecule. Think of iron rusting. Rust is the oxidation of iron – in the presence of oxygen and water, iron molecules will lose electrons over time. Given enough time, this results in the complete break down of objects made of iron.

Rusting iron. Source: Thoughtco
The exact same thing happens in biology. Molecules in your body go through a similar process of oxidation – losing electrons and becoming unstable. This chemical reaction leads to the production of what we call free radicals, which can then go on to damage cells.
What is a free radical?
A free radical is an unstable molecule – unstable because they are missing electrons. They react quickly with other molecules, trying to capture the needed electron to re-gain stability. Free radicals will literally attack the nearest stable molecule, stealing an electron. This leads to the “attacked” molecule becoming a free radical itself, and thus a chain reaction is started. Inside a living cell this can cause terrible damage, ultimately killing the cell.
Antioxidants are the good guys in this situation. They are molecules that neutralize free radicals by donating one of their own electrons. The antioxidant don’t become free radicals by donating an electron because by their very nature they are stable with or without that extra electron.

How free radicals and antioxidants work. Source: h2miraclewater
One of the ways in which DJ-1 responds to oxidative stress is by interacting with different proteins. One of the proteins that DJ-1 has a particular fancy for is called Kelch-like ECH-associated protein 1 (or Keap1). Under normal conditions, Keap1 acts as an anchor to another protein called nuclear factor erythroid-2-related factor 2 (Nrf2). Nrf2 is a major activator protein of antioxidant factors and I have discussed its activities in a previous post (Click here to read that post).
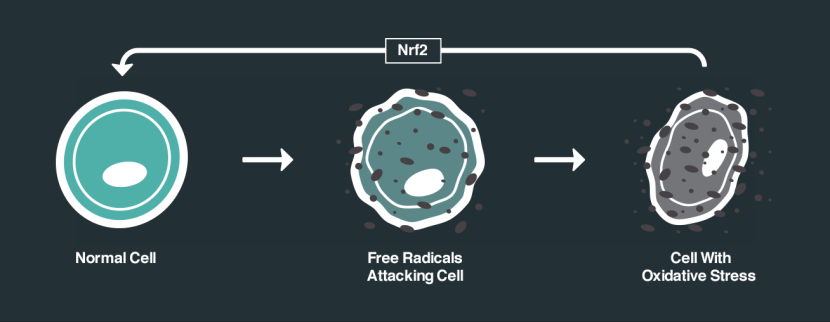
How Nrf2 works. Source: NRF2 science
So without any stress on a cell, Nrf2 is held in place by Keap1. But if oxidative stress suddenly appears, DJ-1 is activated and it grabs Keap1. This results in Nrf2 becoming free to do what it wants, and when Nrf2 is free there is only one thing it wants to do: head for the nucleus of the cell and activate antioxidant factors.
In the nucleus, Nrf2 activates various genes that have antioxidate properties, and by turning on these genes Nrf2 helps to decrease the level of oxidative stress. This antioxidant feature of Nrf2 has a lot of researchers excited and focused on finding drugs that help activate Nrf2.
What else does DJ-1 do?
DJ-1 is also involved in the production of the chemical dopamine. It grabs proteins that block the production of other proteins involved in making dopamine, and when DJ-1 is not functioning properly this means that less dopamine is produced (Click here and here to read more about this).
DJ-1 protein also helps fold newly produced proteins into their proper 3-dimensional shape and also helps re-fold damaged proteins. DJ-1 is known to prevent the clustering (or aggregation) of Parkinson’s associated protein alpha synuclein. Some studies have shown that when the PARK7 gene is mutated, the loss of DJ-1 protein function results in a toxic buildup of misfolded or damaged proteins, eventually leading to cell death.
So as you can see, DJ-1 is a busy little protein with lots of jobs. For a very good review of DJ-1 in Parkinson’s – Click here.
|
# # RECAP #2: DJ-1 is a Parkinson’s-associated protein that has multiple functions within cells. Importantly it binds to a protein called Keap1 and blocks it from inhibiting another protein called Nrf2. This allows Nrf2 to do its job as a major activator of antioxidant factors. DJ-1 also plays a role in the production of the chemical dopamine, which is significantly reduced in Parkinson’s. It also helps to fold proteins properly so that they can do their jobs. # # |
Ok, so what did the researchers find in their new report of DJ-1?
Yes, let’s come back to the new report we are reviewing in today’s post:
 Title: A patient-based model of RNA mis-splicing uncovers treatment targets in Parkinson’s disease.
Title: A patient-based model of RNA mis-splicing uncovers treatment targets in Parkinson’s disease.
Authors: Boussaad I, Obermaier CD, Hanss Z, Bobbili DR, Bolognin S, Glaab E, Wołyńska K, Weisschuh N, De Conti L, May C, Giesert F, Grossmann D, Lambert A, Kirchen S, Biryukov M, Burbulla LF, Massart F, Bohler J, Cruciani G, Schmid B, Kurz-Drexler A, May P, Duga S, Klein C, Schwamborn JC, Marcus K, Woitalla D, Vogt Weisenhorn DM, Wurst W, Baralle M, Krainc D, Gasser T, Wissinger B, Krüger R.
Journal: Sci Transl Med. 2020 Sep 9;12(560):eaau3960.
PMID: 32908004
In this study the researchers were interested in better understanding the biology underlying some of the genetic mutation in the DJ-1 gene that are associated with Parkinson’s. They started their study by looking at levels of DJ-1 protein in cells from people with a specific DJ-1 genetic mutation (the c.192G>C mutation), and they found that the levels of DJ-1 were significantly reduced (compared to unaffected control cells).
In fact, in people with two copies of this particular genetic variation (remember we all have two copies of each gene – one from the mother and one from the father), the level of DJ-1 protein were non-existent. There was literally no DJ-1 protein. Meanwhile, in individuals carrying one copy of the genetic mutation, they produced half of the correct amount of DJ-1 protein.
Further investigations found that these decreases were not due to increased degradation or disposal of the DJ-1 protein, so the investigators assumed that something must be affecting the production of the protein. A deeper analysis of this situation found that the c.192G>C genetic mutation in the DJ-1 gene causes a “skipping” of exon 3.
What??? What does that mean? “Skipping”?
The human DJ-1 gene has 8 exons (Source). As we discussed above, the introns separating each of these exons need to be removed for the proper DJ-1 RNA to be produced.
The researchers found that the G192C point mutation was causing a skipping over the third exon – meaning that exon3 of DJ-1 was missing from the final RNA. This resulted in a significantly shorter mRNA that failed to be translated into protein.
The investigators demonstrated that by genetically engineering a variation of the U1 snRNP component of the spliceosome (we discussed this above), they were able to correct this skipping phenomenon, rescuing both the length of the DJ-1 mRNA and the loss of DJ-1 protein.
Interesting. What did the researchers do next?
Next they turned their attention to potential therapeutic interventions.
Specifically, they looked at the plant cytokinin kinetin (6-furfurylaminopurine).
And what is that?
Kinetin is a cytokinin, which are proteins that stimulate plant growth. Kinetin is used medicinally – people apply it to the skin to treat the effects of aging.
 Kinetin. Source: Wikiwand
Kinetin. Source: Wikiwand
In the current study, however, the researchers wanted to investigare kinetin because it was previously reported to rescue pathologic exon skipping in a fatal inherited condition called familial dysautonomia.
 Source: Familial Dysautonomia Foundation
Source: Familial Dysautonomia Foundation
Familial dysautonomia is a rare fatal autosomal recessive disease, which impairs the development of sensory and autonomic nerves. It is caused by mutations in I-κ-B kinase complex-associated protein (IKBKAP) gene, which lead to splicing abnormalities and results in a deficiency of I-κ-B kinase complex-associated protein (IKAP).
In 2004, a National Institute of Neurological Disorders and Stroke-sponsored drug screening study identified kinetin as a molecule of interest for its ability to rescue the splicing abnormality and restore normal IKAP protein levels in cells that carry the mutation (after just only 1 week of treatment in cell culture!).
This research was followed by a pilot clinical study evaluating kinetin in 8 individuals affected by familial dysautonomia. The results of the study have been published:
 Title: Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia.
Title: Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia.
Authors: Axelrod FB, Liebes L, Gold-Von Simson G, Mendoza S, Mull J, Leyne M, Norcliffe-Kaufmann L, Kaufmann H, Slaugenhaupt SA.
Journal: Pediatr Res. 2011 Nov;70(5):480-3.
PMID: 21775922 (This report is OPEN ACCESS if you would like to read it
This study was the first report of a drug that produced RNA splicing changes in individuals with familial dysautonomia, increasing levels of IKBKAP mRNA in blood cells after just eight days of treatment in 6 of the participants. A remarkable first step towards a treatment.
The researchers investigating the DJ-1 splicing effect were curious to see if kinetin might be able to rescue the reduction in DJ-1 RNA levels that they were observing. They tested a variation of kinetin that is called RECTAS (or RECTifier of Aberrant Splicing)… and guess what?
Yeah, it didn’t really work.
By itself RECTAS did not really increase DJ-1 protein levels in the cells carrying the c.192G>C genetic mutation.
BUT, when the researchers combined RECTAS with phenylbutyric acid – a drug that has previously been reported to increase DJ-1 RNA levels (Click here to read more about this) – they observed an increase in DJ-1 protein. The increase was just 17% of normal control levels after 14 days of treatment, but this was a huge improvement on 0%! And it improved aspects of cellular functioning (such as mitochindrial activity).
More impressively, the researchers grew dopaminergic neurons in cell culture from both normal cells and cells carrying the the c.192G>C DJ-1 genetic mutation. The mutant cells presented fewer dopamine neurons that the control cells (even though there was no difference in total cell count between the two cultures). When the investigators treated the cells with RECTAS and phenylbutyric acid, they saw no difference between the two cell cultures in terms of number of dopamine neurons.
The partial correction of the DJ-1 splicing effect appeared to be sufficient to increase the survival of dopaminergic neurons in that experiment.
|
# # # RECAP #3: A rare mutation in the DJ-1 gene results in a splicing problem that causes reduced amounts of the DJ-1 protein to be produced. Treating cells that carry this genetic mutation with a combination of RECTAS and phenylbutyric acid, partially rescued the effect and improved cellular function. # # # |
Wow! That’s impressive. What did the researchers do next?
So this is the point where this report gets really interesting.
The investigators were intrigued by their discovery, but they were wondering if this splicing issue was just a DJ-1 phenomenon or could other Parkinson’s-associated genes also be affected?
So they turned to the genetic data available on the Michael J. Fox Foundation supported Parkinson’s Progression Markers Initiative (PPMI).
 Started in 2010, PPMI is a major observational clinical study which aims to identify biomarkers of Parkinson’s progression. A biomarker is an objectively measurable physical characteristic associated with a condition. The project has also collected genetic information, and this was what the researchers used to look for other splicing issues.
Started in 2010, PPMI is a major observational clinical study which aims to identify biomarkers of Parkinson’s progression. A biomarker is an objectively measurable physical characteristic associated with a condition. The project has also collected genetic information, and this was what the researchers used to look for other splicing issues.
Amazingly, they found that genetic mutations in U1 binding sites were approximately 40% more common in people with PD than controls on the PPMI database (data from 372 PD cases and 161 controls).
Remember that U1 is one of the five snRNPs that make up the spliceosome (discussed above), and it was the snRNP that the researchers altered to correct the DJ-1 splicing issue.
To double check this results, the investigators repeated our analysis using a larger database of Parkinson’s genetic data, the Parkinson Disease Genome Sequencing Consortium (PDGSC) project, which was established following a National Institute of Neurological Disorders and Stroke workshop in June 2014. The database contains data from 2710 Parkinson’s cases and 5713 controls. In this data set, the genetic mutations in U1 binding sites were only 4% more common in people with PD than controls, but the difference was statistically significant – suggesting that genetic mutations in U1 binding sites are enriched in cases of Parkinson’s.
And this result is particularly interesting because the investigators limited their analysis in these genetic databases to just U1 binding sites. They did not look at other components of the spliceosome (although they are probably having a deep dive right now). This means that genetic variation in other snRNPs could also be potentially influential in Parkinson’s.
Is this splicing issue unique to Parkinson’s? Or are other neurodegenerative conditions also affected?
No.
Splicing effects have been previously associated with Alzheimer’s. For example, this study:
 Title: An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease.
Title: An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease.
Authors: De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van den Bossche T, Cras P, Vandenberghe R, De Deyn PP, Engelborghs S, Van Broeckhoven C, Sleegers K; BELNEU Consortium.
Journal: Acta Neuropathol. 2018 Jun;135(6):827-837.
PMID: 29589097 (This report is OPEN ACCESS if you would like to read it)
In this study, the researchers found three alternative splicing events in the Alzheimer’s-associated gene ABCA7. One of them involved skipping of exon 19 and is abundant in brain tissue.
And other research groups have reported similar exon skipping and splicing-affecting events associated with Alzheimer’s (Click here to read more about this).
So what does it all mean?
Splicing is a critical process in the correct production of protein. Recently researchers have found that a specific genetic mutation in Parkinson’s-associated gene DJ-1 results in a splicing issue that causes reduced levels of DJ-1 protein.
The discovery of this splicing issue not only opens a door to novel factors that could be influencing Parkinson’s, but it also suggests an alternative strategy to “restore cellular abnormalities” and “provides a proof of concept for neuroprotection based on precision medicine strategies” in Parkinson’s (the quote is from the discussion of the report).
This result is really intriguing and I look forward to seeing independent replication of the research, but also further analysis of the concept of mis-splicing within the context of Parkinson’s. It will be interesting to see where this result could potentially lead to with regards to experimental therapies, particularly prodromally (treating Parkinson’s before it is diagnosed).
Lots of food for thought.
 All of the material on this website is licensed under a
All of the material on this website is licensed under a
Creative Commons Attribution 4.0 International License
You can do whatever you like with it!
The banner for today’s post was sourced from Youtube

Love this research. I was at uni getting my BSc in biochemistry when introns were first described. We’ve come a long way. As a biochemist with PD who loves popularization of research (I used to do that for New Scientist), you do a great job.
LikeLike
As always super clear about a subject that is not familiar territory for a lay person. Thank you again for all your superb work.
LikeLike
Thanks for taking the time to make this a bit more understandable. Much appreciated.
LikeLike