|
Here on the SoPD we have discussed the Parkinson’s-associated protein LRRK2 many times. And we look forward to seeing the results of ongoing clinical trials of LRRK2 inhibitors. But there are significant efforts ongoing to develop therapies that can indirectly target dysfunctional LRRK2 pathways (which may help avoid any potential side effects of direct inhibition) Recently, researchers in Scotland and California have published research highlighting one such indriect approach to modulating LRRK2. In today’s post, we will discuss what LRRK2 is, review the new data, and consider the ‘what happens next?’ question.
|
 Prof Dario Alessi. Source: Eureka
Prof Dario Alessi. Source: Eureka
Whenever I read a new research report about the activity of the Parkinson’s-associated protein, LRRK2, my first thought is usually “I wonder what Dario thinks of this?”
And I am not alone in this thought.
Prof Dario Alessi – Director of the Medical Research Council Protein Phosphorylation and Ubiquitylation Unit and Professor of Signal Transduction, at the School of Life Sciences, University of Dundee – is widely recognised as one of the leading experts on the research of this particular protein.
 University of Dundee. Source: Dundee
University of Dundee. Source: Dundee
His thoughts/opinions are widely sought by many in the field – both academic and industry researchers.
And recently his lab – in collaboration with researchers are Stanford University – published a really interesting new report which we will discuss in today’s post.
But first, the obvious question:
What is LRRK2?
Leucine-rich repeat kinase 2 (or LRRK2 – pronounced ‘lark 2’) – also known as ‘Dardarin‘ (from the Basque word “dardara” which means “trembling”) – is an enzyme that has many functions within a cell – from supporting efforts to move things around inside the cell to helping to keep the power on (involved with mitochondrial function).

The many jobs of LRRK2. Source: Researchgate
The LRRK2 gene is made up of many different regions. Each of those regions is involved with the different functions of the eventual protein. As you can see in the image below, the regions of the LRRK2 gene have a variety of different functions:
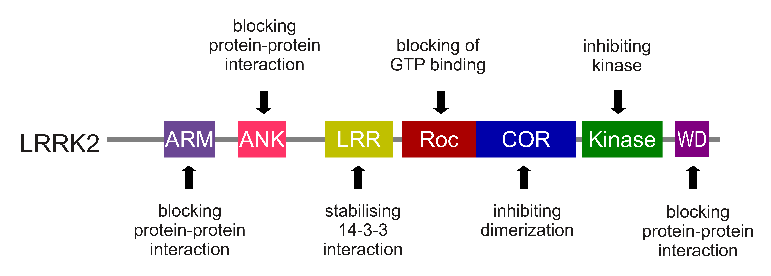
The regions and associated functions of the LRRK2 gene. Source: Intechopen
Genetic errors or variations within the LRRK2 gene are recognised as being some of the most common with regards to increasing ones risk of developing Parkinson’s (LRRK2 variants are present in approximately 1-2% of all cases of Parkinson’s).

The structure of Lrrk2 and where various mutations lie. Source: Intech
As the image above suggests, mutations in the PARK8 gene are also associated with Crohn’s disease (Click here and here for more on this) – though that mutation is in a different location to those associated with Parkinson’s. And one particularly common Parkinson’s-associated LRRK2 mutation – called G2019S – is also associated with increased risk of certain types of cancer, especially for hormone-related cancer and breast cancer in women – Click here to read more about this. If you have a G2019S mutation, no reason to panic – but it is good to be aware of this association and have regular check ups.
The G2019S mutation (the name designates its location on the gene) is the most common LRRK2 mutation. In some populations of people it can be found in 40% of people with Parkinson’s (Click here to read more about this). But what is interesting about this mutation is that it gives rise to a LRRK2 enzyme that is hyperactive.
 LRRK2 protein. Source: Youtube
LRRK2 protein. Source: Youtube
As a protein, LRRK2 interacts with many different types of other proteins, and you can imagine that in a finely balanced environment like the cells that a mutant ‘hyperactive’ form of LRRK2 is going to cause problems. The consequences of this constantly active form of LRRK2 protein is believed to be influential in the cell death in LRRK2-associated Parkinson’s.
This has led to the development of treatments that inhibit the hyperactive version of LRRK2. Some of these treatments are now being clinically tested, and leading the pack here is a biotech company called Denali Therapeutics.
Founded in 2013, by a group of former Genentech executives, this San Francisco-based biotech company now has two drugs – LRRK2 inhibitors called DNL-201 and DNL-151 – in Phase I safety/tolerability clinical testing for Parkinson’s (Click here and here to read more about these trials).
In addition, a biotech company is called Ionis Pharmaceuticals is developing a novel LRRK2 approach.
Previously known as Isis Pharmaceuticals (until it changed this name in December 2015 when the label ISIS took on negative connotations), Ionis is collaborating with the pharmaceutical company Biogen to target LRRK2 in Parkinson’s.
The two companies have recently registered a Phase I clinical trial to evaluate their lead candidate, called BIIB094. BIIB094 is an antisense oligonucleotide, which is a method of blocking LRRK2 RNA before it can be used to make LRRK2 protein (we have discussed antisense oligonucleotides in a previous SoPD post – click here to read that post). The trial is a Phase I clinical trial to evaluate the safety and tolerability of single and multiple doses of BIIB094 (Click here to learn more about that trial).
|
RECAP #1: LRRK2 is a Parkinson’s-associated gene (functional region of DNA). The LRRK2 gene provides instructions for producing a multi-functional protein. When mutated, the gene provides instructions for a hyperactive form of the LRRK2 protein, which is believed to disrupt normal cellular function. Multiple biotech companies are developing therapies that inhibit the hyperactive form of the LRRK2 protein.
|
One of the potential issues with LRRK2 inhibitors and inhibition, however, is the possibility of ‘off target effects’. LRRK2 is a busy protein with lots of different functions and inhibiting some of those functions could lead to undesired side effects. I am speculating here, we have no evidence in human to suggest that this occurs, but it has led to researchers looking for alternative ways of suppressing LRRK2 activity.
Rather than directly interacting with the LRRK2 protein and inhibiting it, scientists have been asking if there are there better ways to reduce its activity?
And? Are there?
This is where we return back to Prof Alessi, who I mentioned in the intro.
Recently Prof Alessi and Prof Suzanne Pfeffer of Standford University have been collaborating on an effort to identify alternative methods of modulating LRRK2 activity, and in October they published a report which points towards one potential approach.
This is the report:
 Title: PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins.
Title: PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins.
Authors: Berndsen K, Lis P, Yeshaw WM, Wawro PS, Nirujogi RS, Wightman M, Macartney T, Dorward M, Knebel A, Tonelli F, Pfeffer SR, Alessi DR.
Journal: Elife. 2019 Oct 30;8. pii: e50416.
PMID: 31663853 (This report is OPEN ACCESS if you would like to read it)
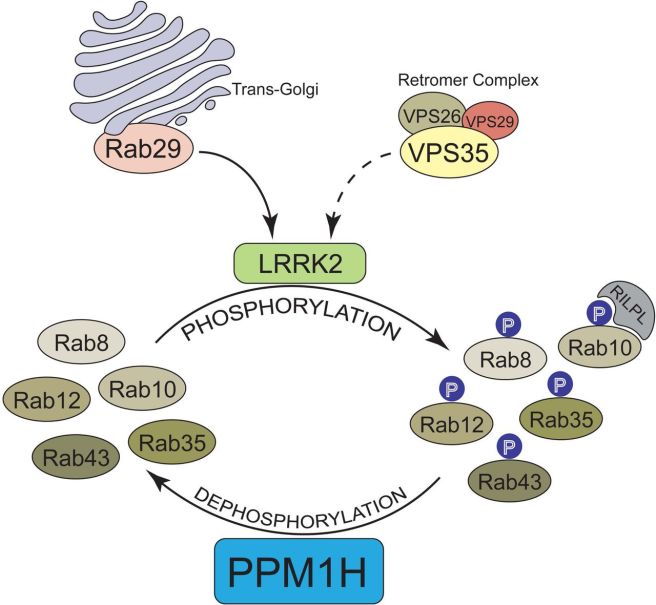
In this study, the researchers were interested in a group of proteins that LRRK2 interacts with, which are called RAB proteins.
What are RAB proteins?
RAB proteins are a group of proteins that sit on the inside of cell membranes. Each RAB protein has unique subcellular membrane distribution and they take part in various functions. RAB proteins form the largest branch of a large family of proteins called GTPases.
What are GTPases?
These proteins regulate the transportation of vesicles (the small bags of necessary proteins that live within a cell) between different membranes within cells. In the basic image below, you can see an examples of how Rab proteins (green squares) are involved with transporting vesicles.
Like other GTPases, the RAB proteins have two conformations: an inactive form which is maintained by being bound to a protein called guanosine diphosphate (or GDP) and an active form, which can be identified by being bound to guanosine triphosphate (or GTP).The switching between states occurs through the process of phosphorylation.
Remind me again: What is phos…phoryl…ation?
Phosphorylation of a protein is basically the process of turning it on or off – making it useful or inactivating it. From allowing a protein to fold in a particular manner to actually activating/deactivating the function of a protein, phosphorylation is a critical function in cellular biology.

Phosphorylation of a kinase protein. Source: Nature
Phosphorylation occurs via the addition or removal of phosphates. GDP and GTP (mentioned above) are both types of phosphates. Their addition or removal determines the state of the RAB proteins, it is controlled by proteins called phosphatases.
Understand that LRRK2 actively phosphorylates certain RAB proteins (Click here for a thorough review on this topic). By phosphorylating RAB proteins, LRRK2 blocks the ability of the RAB proteins to interact with other proteins, thereby trapping the phosphorylated RAB protein on the membrane and unable to performing its task.
|
RECAP #2: RAB proteins are membrane proteins that are involved with different cellular activities (many involving the movement of other proteins). Some RAB proteins are phosphorylated (activated) by LRRK2. Since RAB protein activities are affected by LRRK2, they represent an indirect method of therapeutically regulating LRRK2 activity (rather than blocking LRRK2 directly). The issue is identifying phosphotases (proteins that activate/inactivate proteins) that counter LRRK2 regulation of RAB proteins.
|
In their new study, the Prof Alessi and colleagues wanted to identify phosophatases that dephosphorylate (or remove phosphates from) one particular RAB protein: RAB10.
Why?
LRRK2 phosphorylates RAB proteins, but there are other phosphatases that dephosphorylate RAB proteins. LRRK2 and these other phosphatases work in opposition to each other – phosphorylating and dephosphorylating proteins, respectively.
So, by identifying phosphatases that dephosphorylate RAB10, we may be able to develop a useful way of countering LRRK2 activity, which could have therapeutic potential.
The hope is that while inhibiting LRRK2 may have undesirable side effects that need to be carefully controlled, going after a downstream protein like RAB10 could be a more attractive treatment option.
Ok, but why RAB10 in particular?
So in late 2018, Prof Pfeffer and Prof Alessi published this report:
 Title: A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain.
Title: A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain.
Authors: Dhekne HS, Yanatori I, Gomez RC, Tonelli F, Diez F, Schüle B, Steger M, Alessi DR, Pfeffer SR.
Journal: Elife. 2018 Nov 6;7. pii: e40202.
PMID: 30398148 (This report is OPEN ACCESS if you would like to read it)
In this study, they reported that Rab10 phosphorylation blocks a neuroprotective pathway involving a protein called sonic hedgehog (we reviewed this report in a previous SoPD post – Click here to read that post).
You mean like the video game character?
Yep. Genetisists have produced some wonderful names for genes/proteins (“lunatic fringe” and “mothers against decapentaplegia (MAD)” are two of my favourites).
They found that when LRRK2 is mutated and becomes hyperactive, it results in increased levels phosphorylated RAB10. Increased levels of phosphorylated RAB10 blocks a sonic hedghog neuroprotective pathway.
Since publishing that report, the research team has been trying to identify phosophatases that will dephosphorylate RAB10. If RAB10 specific phosophatases can be identified, perhaps drugs could be designed that will act like them. Such a drug could be potentially useful in the treatment of LRRK2-associated Parkinson’s.
And their search for phosophatases that will dephosphorylate RAB10 has given rise to the report we are reviewing in today’s post.
I see. So have they identified any RAB10 specific phosophatases?
First, they wanted to evaluate whether two particular phosophatases – PP1 (Protein Phosphatase-1) or PP2A (Protein Phosphatase-2A) – dephosphorylate RAB proteins. Both have previously been reported to regulate LRRK2 activity (Click here and here to read more about this).
The researchers found that PP1 and PP2A do not control RAB phosphorylation.
This result required the researchers to begin a hunt for phosphotases that do dephosphorylate RAB proteins.
|
RECAP #3: LRRK2 phosphorylates RAB10, but too much phosphorylated RAB10 blocks a neuroprotective pathway which may be important in Parkinson’s. The researchers hoped to identify phosphotases that will dephosphorylate RAB10 and counter the effect of dysfunctional LRRK2 activity.
|
And did they have any luck with their hunt for phosphotases that dephosphorylate RAB proteins?
They used three different screens and the top protein across two of them was a protein called PPM1H (or Metal-dependent protein phosphatases [PPM] family serine/threonine phosphatase). Before this study very little was known about PPM1H, but the researchers found that in cells in culture it potently inhibits LRRK2-mediated RAB10 protein phosphorylation.
Interestingly, PPM1H also dephosphorylated other RAB proteins (such as RAB8A and RAB35).
The researchers noted that it would be interesting “to explore whether increased expression or activity of PPM enzymes protects patients with LRRK2 mutations from developing Parkinson’s by enhancing RAB protein dephosphorylation“. But they also raised the intriguing question of whether “reduced expression or activity of PPM1H phosphatase would be expected to promote RAB protein phosphorylation” and thus enhance the risk of developing Parkinson’s.
Obviously further research is required in this area, but PPM1H may represent a novel mechanism by which LRRK2-associated Parkinson’s could be treated in the future.
So what does it all mean?
Regular readers of this blog will have noticed that there have been a number of post recently regarding this Parkinson’s-associated protein called LRRK2. There is an absolute tidal wave research coming down the pipe that is focused on this key player in Parkinson’s.
And it is fair to say that one of the reasons for this flood of data is the dedicated contribution of Google co-founder Sergey Brin and his family foundation. The Brin family have been affected by Parkinson’s (Sergey, his mother and his aunt all carry a LRRK2-G2019S genetic variant).
 The Brin Family – Sergey and his mother on the right. Source: CS
The Brin Family – Sergey and his mother on the right. Source: CS
Sergey may never go on to develop the condition, but he has obviously decided not to take any chances, and he has taken out an “insurance policy” by investing hundreds of millions of dollars into Parkinson’s research.
Researchers like Prof Alessi in Dundee (Scotland) and Prof

All of the material on this website is licensed under a
Creative Commons Attribution 4.0 International License
You can do whatever you like with it!
The banner for today’s post was sourced from Wikipedia





3 thoughts on “The Lords of LRRK”